Abstract
Introduction: Clonal hematopoiesis is an increasingly well-established risk factor for developing therapy-related acute myeloid leukemia and myelodysplastic syndrome (t-AML/t-MDS), particularly in patients receiving chemotherapy. In some cases, leukemia-associated mutations can be detected at low frequencies in prior samples, and the same mutation is expanded significantly in t-AML/t-MDS samples. However, most of these studies have been conducted using whole mononuclear cells populations and it is unclear whether these mutations reside in the stem cell compartment. Here, we present DNA sequencing results from sorted mobilized stem cells from patients with multiple myeloma (MM) treated with autologous hematopoietic stem cell transplant (AHSCT) who subsequently developed t-AML/t-MDS.
Methods: Patients with MM treated with AHSCT at the University of Arkansas for Medical Sciences Hospital who subsequently developed t-AML/t-MDS were identified. Clinical data, including the results of commercially available targeted gene sequencing, was recorded and unused stem cell collection specimens were retrieved. Specimens were sorted by flow cytometry into subpopulations, including normal hematopoietic stem cells (nlHSCs, Lin-/CD34+/CD38-), aberrant hematopoietic stem cells (Ab-HSC, defined as phenotypic HSCs that are additionally either CD99+ or CD123+ or IL1RAP+), megakaryocyte-erythroid progenitors (MEPs), granulocyte-macrophage progenitors (GMPs), granulocytes (Grans), T-cells, and B-cells. DNA was extracted and targeted deep sequencing was performed by NGS (Genoptix). The low frequency variants (<1%) are presently being validated by independent high sensitivity assays. If available, DNA from CD138+ myeloma cells collected at the time of initial diagnosis was sequenced. The libraries were then queried for mutations identified by commercially available targeted sequencing performed on t-AML/t-MDS samples, after the exclusion of germline mutations. The protocol was approved by the IRBs at the participating institutions.
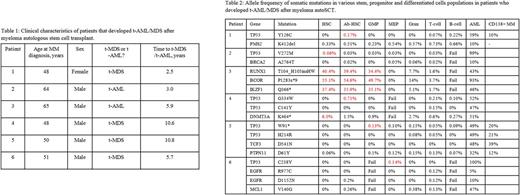
Results: Six patients were identified. All had unused stem cell collection samples and commercially available DNA sequencing performed at the time of t-AML/t-MDS diagnosis. Two had DNA from CD138+ MM cells available for sequencing. Clinical characteristics of these patients are shown in Table 1. The results of mutational frequencies among the subpopulations are show in in Table 2. In all patients, mutations subsequently found in t-AML/t-MDS samples were found among at least one stem or progenitor subpopulation. Variant allele frequency ranged from 0.02%- 55.1%. The low frequency variants are presently being validated by independent assays. Mutations were particularly enriched in nlHSCs or Ab-HSCs in 4 out of 6 patients and were enriched in phenotypic GMP or MEP progenitors in the other two cases. In particular, Patient 3 was found to have high frequencies of RUNX1, IKZF1, and BCOR mutations in nlHSC, Ab-HSC, and GMP populations, with much lower frequencies in the mature populations, and with none in the MEP population. In both patients with evaluable CD138+ MM samples, the same TP53 mutations present in the MM were also present in the HSCs as well as mature B lymphocytes, demonstrating persistence during plasma cell differentiation. .
Conclusions: Our results show for the first time the presence and distribution of t-AML/t-MDS-associated mutations in patients at the time of MM treatment, many years before eventual transformation. While mutations to tumor suppressor genes such as TP53 have previously been detected at low frequency in unsorted antecedent samples, we show that these mutations preferentially reside in nlHSCs, Ab-HSCs, and immature progenitor populations which enrich over time, leading to high frequencies at the time of t-AML/t-MDS diagnosis. Finally, in 2 of 2 evaluable patients, we demonstrate the presence of the same tumor initiating mutation in both CD138+ MM and t-MDS samples, suggesting that common ancestral tumor initiating cells harboring these mutations may have given rise to both the MM and t-MDS.
Barlogie: Millenium Pharmaceuticals: Consultancy, Research Funding; Celgene Corporation: Consultancy, Research Funding. Morgan: Takeda: Consultancy, Honoraria; Celgene: Consultancy, Honoraria, Research Funding; Bristol Myers: Consultancy, Honoraria. Steidl: Bayer Healthcare: Consultancy; Novartis: Research Funding; Celgene: Consultancy; GlaxoSmithKline: Research Funding; Aileron Therapeutics: Consultancy, Research Funding. Will: Novartis Pharmaceuticals: Consultancy, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal